
Clinicopathological characterization of next-generation sequencing detected mutations in lung cancers—a single-center experience
Highlight box
Key findings
• Next-generation sequencing (NGS) results differ regarding clinicopathological patient characteristics in non-small cell lung cancer (NSCLC).
What is known and what is new?
• NGS panel testing is mandatory in many NSCLC cases to detect even rare driver mutations.
• Low amount of material or lack of reimbursement prevents complete panel testing in many cases.
What is the implication, and what should change now?
• Increasing pre-test probability by selecting patients based on clinicopathological characteristics may help to provide adequate testing, wherever resources are scarce.
Introduction
Non-small cell lung cancer (NSCLC) is still the leading cause of cancer-related deaths worldwide (1). However, for a proportion of patients with activating driver mutations, development of targeted therapies has led to dramatically improved outcome (2,3). Despite many advances in molecular procedures many NSCLC patients do not receive full panel testing due to limited amount of material, lacking reimbursement of next-generation sequencing (NGS) or other reasons (4).
In cases of EGFR mutations or fusions of the ALK, several clinicopathological features were shown to characterize patients with a higher pre-test probability for these alterations. In particular, younger females with no history of cigarette smoking have been found to harbor such mutations (5,6). Broad molecular multi-gene testing became especially important since more and more targeted drugs for in part rare mutations had been approved or are in advanced development (7).
Furthermore, not only metastasized patients but also patients with local disease receive molecular testing as the anti-EGFR targeted osimertinib has been approved for adjuvant treatment (8). Currently, there is limited understanding of how molecular profiles differ in patients with early-stage disease compared to advanced stages. In addition, while financial reimbursement for NGS testing is available in individual countries, this option is not available worldwide. Therefore, it would be beneficial to choose different patterns of diagnostic in distinct cases to spare tumor tissue, money, and time.
For this reason, it would be important to better characterize clinicopathological features of patients with distinct mutations. This might help to improve pre-test probability in cases with limited tumor material or in patients without indication for routine clinical testing e.g., due to histology or tumor stage.
In this study, we sought to correlate different clinicopathological features such as histology, tumor stage, PD-L1 expression, and patients’ baseline characteristics with the results of NGS testing. We present this article in accordance with the STROBE reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-751/rc).
Methods
Patient population and study design
In this retrospective cross-sectional analysis, we used data from patients with a confirmed NSCLC diagnosis treated at the University Hospital, LMU Munich. We included all patients referred to NGS testing by the thoracic oncology department between 2018 and 2021. We included patients with both, non-squamous cell carcinoma (SCC), and SCC. To analyze patients’ baseline characteristics and clinicopathological features, we collected data on sex, age, survival status and overall survival (OS), Eastern Cooperative Oncology Group (ECOG) status, histology, tumor stage, PD-L1 tumor proportional score, as well as results of the NGS testing. We categorized stage into patients with metastases (stage IV) and without metastases (stage I–III). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval for this study was obtained from the ethics board of the Ludwig-Maximilians University Munich (reference number 474-16 UE). Due to the retrospective nature of the data, the ethics board consented to the analysis without requirements beyond the anonymization of the data prior to the analysis. Individual consent for this retrospective analysis was waived.
NGS testing
All patients received NGS panel testing at the Institute of Pathology of the University of Munich (LMU). Panel-guided NGS as well as variant calling was performed as described in detail previously (9). Briefly, nucleic acids were extracted from formalin-fixed paraffin-embedded (FFPE) tissue sections using the GeneRead (DNA) and RNeasy FFPE kits (RNA) (both from Qiagen, Hilden, Germany). NGS was performed on an Ion Torrent GeneStudio S5 Prime (Thermo Fisher, Waltham, MA, USA) or Illumina 500 Next Seq (Illlumina, San Diego, CA, USA) systems. The Oncomine Focus Panel (covering 52 cancer-associated genes) until November 2018 followed by the Oncomine Comprehensive v.3 assay [limited to the predefined 20 genes of the national Network Genomic Medicine lung cancer (nNGM)] till January 2020 (both Thermo Fisher). In January 2020 the predefined genes list was expanded to 26 genes (nNGM v2). The FusionPlex Lung panel (ArcherDX, Boulder, CO, USA) replaced the Oncomine fusion analysis part in September 2020. Since March 2021 a custom VariantPlex panel, covering the 26 nNGM v2 genes, and the FusionPlex Lung panel (both ArcherDX/Invitae) were applied. Moreover, certain cases were analyzed with the AmpliSeq Colon and Lung Cancer v2 DNA only panel (22 genes, Thermo Fisher).
Single-nucleotide variants (SNVs), multi-nucleotide variants (MNVs), small insertions, deletions, indels, copy number variation (CNV), and gene fusions were analyzed. Results were extracted from the pathology reports for further clinicopathological analyses.
As mentioned above, during the observation period due to technological and therapeutical progress the panels used for testing included an increasing number of genes. To reflect this issue when calculating the prevalence of genes not included in all panels we analyzed following mutations in subgroups of patients: RET (n=133 patients), PTEN and TP53 (n=102 patients), STK11 (n=86 patients), KIT and PDGFRA (n=59 patients), and AR, CCND1, ERG, MYC, and MYCN (n=58 patients).
Analysis of all mutations
For the comparison of baseline characteristics and clinicopathological features we categorized patients into patients with and without any mutation, irrespective of whether patients had one or more mutations. In the analysis of the frequency of distinct mutations and the stratification by sex, as well as metastases status and histology, the overall n refers to the number of mutations and not the number of patients.
Analysis of five most common mutations and number of mutations
For a more thorough investigation of differences in baseline characteristics and clinicopathological features we selected the five most common mutations, and categorized patients according to whether they had one of these five mutations (one category for each mutation), or no mutations at all. Additionally, we categorized patients according to the number of different gene mutations they had into no mutations, one, two, and three or more mutations.
Statistical analysis
Metric variables were presented as means with standard deviation (SD) and compared using t-test in the two-group comparison, and analysis of variance (ANOVA) in the comparison of more than two groups. Categorical variables were reported as absolute and relative frequencies and compared between groups using χ2-test and Fisher’s exact test (cell numbers <6). To calculate the relative frequency of genes not included in all panels we used the number of patients in subgroups mentioned above. We used Kaplan Meier curves, with log-rank test to compare survival between groups. Statistical significance was determined using two-sided P values with alpha errors <0.05. Data analysis was performed using R version 4.0.0 and RStudio version 1.4. Tables and figures were created in RStudio and Microsoft Excel.
Results
Patient characteristics stratified by mutation status and histological type
Overall, 154 matched the inclusion criteria for this study. Of those, NGS testing found any mutations in 108 (70.1%) patients and no mutation in 46 (29.9%) patients. Mean age overall was 62.7 (SD =12.5), and 54.5% of patients were male. The proportion of males in patients without mutations was higher than in the group with mutations (60.9% vs. 51.9%); however, this difference was not statistically significant. The majority of patients tested presented with adenocarcinoma (AC) (79.2%) at time of diagnosis 64.9% were in stage IV, 18.8% in stage III, and 16.2% in stage I and II. We found that patients/tumors with mutations had significantly higher levels of PD-L1 expression compared to patients/tumors without mutations (36.4% vs. 19.2%, P=0.005). No other baseline characteristics or clinicopathological features differed significantly between patients with and without mutations. In patients diagnosed with AC mean age was lower (61.8 vs. 66.3 years, P=0.07), mean PD-L1 expression was higher (33.6% vs. 22.7%, P=0.01), and female sex was more common (49.2% vs. 31.3%, P=0.11) compared to patients with SCC. Distribution of stage at diagnosis was significantly different in patients with AC and SCC (P<0.001). While the majority of patients with AC were diagnosed in stage IV (73.0%) only 34.4% with SCC had stage IV disease. Further, 10.7% of patients with AC were diagnosed in stage I and II, and 16.4% in stage III, while this distribution was 37.5% and 28.1% in patients SCC. We displayed results for all characteristics and features overall and stratified by mutations status in Table 1.
Table 1
| Characteristics | All patients (n=154) | With mutation (n=108) | Without mutation (n=46) | P value |
|---|---|---|---|---|
| Age (years), mean ± SD | 62.7±12.5 | 62.5±13.0 | 63.2±11.5 | 0.73 |
| Sex, n (%) | 0.39 | |||
| Male | 84 (54.5) | 56 (51.9) | 28 (60.9) | |
| Female | 70 (45.5) | 52 (48.1) | 18 (39.1) | |
| Histology, n (%) | 0.98 | |||
| AC | 122 (79.2) | 85 (78.7) | 37 (80.4) | |
| SCC | 32 (20.8) | 23 (21.3) | 9 (19.6) | |
| Stage at diagnosis, n (%) | 0.70 | |||
| I and II | 25 (16.2) | 19 (17.6) | 6 (13.0) | |
| III | 29 (18.8) | 19 (17.6) | 10 (21.7) | |
| IV | 100 (64.9) | 69 (63.9) | 31 (67.4) | |
| PD-L1 status (%), mean ± SD | 31.2±36.1 | 36.4±37.9 | 19.2±28.6 | 0.005 |
| PD-L1 status, n (%) | ||||
| <1% | 45 (29.2) | 30 (27.8) | 15 (32.6) | 0.68 |
| 1% to 50% | 46 (29.9) | 28 (25.9) | 18 (39.1) | 0.14 |
| >50% | 46 (29.9) | 38 (35.2) | 8 (17.4) | 0.04 |
| Missing | 17 (11.0) | 12 (11.1) | 5 (10.9) | – |
| ECOG, n (%) | ||||
| 0 | 69 (44.8) | 46 (42.6) | 23 (50.0) | >0.99 |
| 1 | 23 (14.9) | 16 (14.8) | 7 (15.2) | 0.93 |
| 2 | 7 (4.5) | 4 (3.7) | 3 (6.5) | 0.68 |
| Not available | 55 (35.7) | 42 (38.9) | 13 (28.3) | – |
Baseline characteristics and clinicopathological features of patient population overall and stratified by mutation status (no mutation detected vs. any mutation). Metric variables reported as means with SD with P values from t-test. Categorical variables reported as absolute and relative frequencies with P value from χ2-test and Fisher’s exact test (cell number <6). SD, standard deviation; AC, adenocarcinoma; SCC, squamous cell carcinoma; ECOG, Eastern Cooperative Oncology Group.
Analysis of distinct mutations
The five most common mutations found in the total cohort were TP53 (n=39, 25.3%), KRAS (n=30, 19.5%), EGFR (n=24, 15.6%), MET (n=17, 11.0%) with MET skipping exon 14 in nine of these, and PIK3CA (n=10, 6.5%). The distribution of EGFR mutation was significantly different between males and female (9.5% vs. 22.9%, P=0.04). Relative frequencies overall and by sex for all mutations can be found in Figure 1.
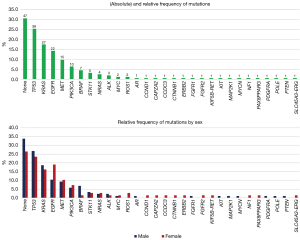
Distribution of mutations dependent on stage at diagnosis are displayed in Figure 2. Although the distribution was quite similar, there were some notable differences regarding stage. The proportion of patients with EGFR mutation was higher in patients with stage IV with 18.0% compared to stage III with 10.3% and stage I and II with 12.0% (P=0.52). Whereas the proportion of patients with PIK3CA (5.0% vs. 6.9% vs. 12.0%, P=0.44), and BRAF (3.0% vs. 6.9% vs. 12.0%, P=0.17) was lower in patients with stage IV, III and I and II, respectively.

Figure 3 displays the distribution of distinct mutations in AC and SCC. The most common mutations in AC were TP53 (37.7%), KRAS (21.3%), EGFR (16.4%), MET (13.1%), and PIK3CA (2.5%). In general, the variety of different mutations was higher in AC, where 22 different mutations were detected. In SCC, 13 different mutations were detected. The most common ones were TP53 (38.5%), PIK3CA (21.9%), KRAS (12.5%), EGFR (12.5%), and MET (3.1%). The difference between the prevalence of PIK3CA in AC and SCC was significant (P<0.001).

Analysis of characteristics of patients with five most common mutations
Of all patients with one of the five most common mutations patients with EGFR mutations were the youngest (57.0 years) and patients with MET alterations were the oldest (66.4 years). The proportion of males was lowest in EGFR (33.3%) and highest in TP53 (59.0%). Mean values of PD-L1 expressions varied from 24.0% in EGFR, around 40.0% in PIK3CA, KRAS, and TP53, up to 56.8% in patients with MET alterations.
Table 2 displays all results from the analysis of the five most common mutations. As patients can have more than one mutation, no significance tests were performed in this part of the analysis.
Table 2
| Characteristics | No mutation (n=46) | EGFR (n=24) | KRAS (n=30) | MET (n=17) | PIK3CA (n=10) | TP35 (n=39) |
|---|---|---|---|---|---|---|
| Age (years), mean ± SD | 63.2±11.5 | 57.0±13.6 | 64.7±11.0 | 66.4±8.9 | 65.7±10.8 | 62.4±14.1 |
| Age (years), n (%) | ||||||
| <60 | 17 (37.0) | 14 (58.3) | 7 (23.3) | 4 (23.5) | 3 (30.0) | 15 (38.5) |
| 60–75 | 22 (47.8) | 9 (37.5) | 19 (63.3) | 10 (58.8) | 6 (60.0) | 17 (43.6) |
| >75 | 7 (15.2) | 1 (4.2) | 4 (13.3) | 3 (17.6) | 1 (10.0) | 7 (17.9) |
| Gender, n (%) | ||||||
| Male | 28 (60.9) | 8 (33.3) | 16 (53.3) | 8 (47.1) | 5 (50.0) | 23 (59.0) |
| Female | 18 (39.1) | 16 (66.7) | 14 (46.7) | 9 (52.9) | 5 (50.0) | 16 (41.0) |
| PD-L1 status (%), mean ± SD | 19.2±28.6 | 24.0±31.9 | 41.1±37.9 | 56.8±35.1 | 37.1±38.5 | 40.0±38.8 |
| PD-L1 status, n (%) | ||||||
| <1% | 15 (32.6) | 9 (37.5) | 8 (26.7) | 1 (5.9) | 2 (20.0) | 13 (33.3) |
| 1% to 50% | 18 (39.1) | 5 (20.8) | 13 (43.3) | 12 (70.6) | 4 (40.0) | 8 (20.5) |
| >50% | 8 (17.4) | 4 (16.7) | 7 (23.3) | 4 (23.5) | 4 (40.0) | 15 (38.5) |
| Missing | 5 (10.9) | 6 (25.0) | 2 (6.7) | 0 (0.0) | 0 (0.0) | 3 (7.7) |
| ECOG, n (%) | ||||||
| 0 | 23 (50.0) | 12 (50.0) | 13 (43.3) | 7 (41.2) | 5 (50.0) | 15 (38.5) |
| 1 | 7 (15.2) | 3 (12.5) | 1 (3.3) | 2 (11.8) | 2 (20.0) | 4 (10.3) |
| 2 | 3 (6.5) | 0 (0.0) | 2 (6.7) | 1 (5.9) | 0 (0.0) | 1 (2.6) |
| Not available | 13 (28.3) | 9 (37.5) | 14 (46.7) | 7 (41.2) | 3 (30.0) | 19 (48.7) |
Comparison of baseline characteristics and clinicopathological features of patients with the five most common mutations. Group no mutation refers to patients without any detected mutation. Metric variables reported as means with SD with P values from t-test. Categorical variables reported as absolute and relative frequencies with P value from χ2-test and Fisher’s exact test (cell number <6). SD, standard deviation; ECOG, Eastern Cooperative Oncology Group.
Analysis of characteristics stratified by number of mutations
In total, 40.9% (n=63) of patients had only one detected mutation, 21.4% (n=33) had two different mutations, and 7.1% of patients were found to have three or more different mutations. The proportion of female patients increased with increasing number of mutations from 40.4% in patients without mutation to 54.5% in patients with three or more mutations, however this increase was not significant. Mean PD-L1 expression increased from 19.2% in patients without mutations to 43.0% in patients with three or more mutations (P=0.07). Table 3 displays all characteristics stratified by number of mutations.
Table 3
| Characteristics | No mutation (n=47) |
One mutation (n=63) |
Two mutations (n=33) |
Three or more mutations (n=11) |
P value |
|---|---|---|---|---|---|
| Age (years), mean ± SD | 63.1±11.4 | 62.6±13.8 | 62.9±13.1 | 60.7±7.54 | 0.96 |
| Sex, n (%) | 0.81 | ||||
| Male | 28 (59.6) | 34 (54.0) | 17 (51.5) | 5 (45.5) | |
| Female | 19 (40.4) | 29 (46.0) | 16 (48.5) | 6 (54.5) | |
| Histological type, n (%) | 0.18 | ||||
| AC | 38 (80.9) | 46 (73.0) | 30 (90.9) | 8 (72.7) | |
| SCC | 9 (19.1) | 17 (27.0) | 3 (9.1) | 3 (27.3) | |
| Stage at diagnosis, n (%) | 0.86 | ||||
| I and II | 6 (12.8) | 11 (17.5) | 7 (21.2) | 1 (9.1) | |
| III | 10 (21.3) | 12 (19.0) | 4 (12.1) | 3 (27.3) | |
| IV | 31 (66.0) | 40 (63.5) | 22 (66.7) | 7 (63.6) | |
| PD-L1 status (%), mean ± SD | 19.2±28.6 | 35.7±38.8 | 35.3±35.7 | 43.0±42.4 | 0.07 |
| PD-L1 status, n (%) | |||||
| <1% | 15 (31.9) | 16 (25.4) | 11 (33.3) | 3 (27.3) | 0.86 |
| 1% to 50% | 18 (38.3) | 17 (27.0) | 8 (24.2) | 3 (27.3) | 0.41 |
| >50% | 8 (17.0) | 22 (34.9) | 12 (36.4) | 4 (36.4) | 0.14 |
| Missing | 6 (12.8) | 8 (12.7) | 2 (6.1) | 1 (9.1) | – |
| ECOG, n (%) | |||||
| 0 | 23 (48.9) | 30 (47.6) | 11 (33.3) | 5 (45.5) | 0.76 |
| 1 | 7 (14.9) | 14 (22.2) | 1 (3.0) | 1 (9.1) | 0.33 |
| 2 | 3 (6.4) | 2 (3.2) | 2 (6.1) | 0 (0.0) | 0.48 |
| Not available | 14 (29.8) | 17 (27.0) | 19 (57.6) | 5 (45.5) | – |
Comparison of baseline characteristics and clinicopathological features stratified by number of mutations. Group no mutation refers to patients without any detected mutation. Metric variables reported as means with SD with P values from t-test. Categorical variables reported as absolute and relative frequencies with P value from χ2-test and Fisher’s exact test (cell number <6). SD, standard deviation; AC, adenocarcinoma; SCC, squamous cell carcinoma; ECOG, Eastern Cooperative Oncology Group.
Survival analysis
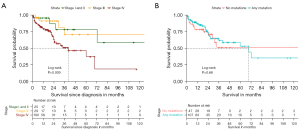
We compared OS of different subgroups in our cohort. As expected, patients with stage IV disease had significantly reduced OS compared to patients with localized or local advanced disease (log-rank P=0.009, Figure 4A). However, we could not detect a significant difference in survival between patients with or without any mutation (Figure 4B). There was also no OS benefit for one distinct mutation (data not shown), when stratified for the five most common mutations.
Discussion
In this retrospective, single center analysis, we sought to evaluate the clinicopathological characteristics of different oncogenic driver mutations in NSCLC patients.
We found a significant association of PD-L1 expression and presence of driver mutations. Overall mean PD-L1 expression was higher in patients with mutations compared to patients without a mutation (P=0.005) and the proportion of patients with a PD-L1 expression greater than 50% was significantly higher in the presence of a driver mutation. Patients with MET alterations, those with PIK3CA mutations or those with three or more mutations showed the highest levels of PD-L1 expression. This is confirmed by recent investigations showing that MET and PIK3CA alterations in NSCLCs are significantly related to high PD-L1 expression (10,11). On the other hand, patients harboring an EGFR mutation had only a slight increase of PD-L1 levels compared to patients with no mutation. Levels of PD-L1 expression are in line with a current publication of Li and colleagues, showing that tumors harboring EGFR mutations had a PD-L1 expression in 18.8% of cases and that PIK3CA, MET, and KRAS mutations were associated with higher PD-L1 expression (11). In contrast to our cohort, this work showed that overall PD-L1 expression was higher in patients without any mutations. This may be due to differing distribution of driver mutations in Asian compared to European populations. However, another meta-analysis revealed a positive association between EGFR mutations and PD-L1 expression (12), underlining the results of our study.
We found differences in sex specific distribution of distinct mutations. TP53, and BRAF mutations were more frequent in male patients, whereas EGFR (P=0.04) and AR alterations were more common in females. Interestingly, the same association was found in a current publication investigating a set of 1,017 lung cancer cases (13). These observations may help inform further studies of sex differences in carcinogenesis and cancer risk. For instance, the higher proportion of TP53 and BRAF mutations in male patients might be explained by a higher smoking history in men. In addition, there may be a role for individualized mutation specific diagnostics in small biopsies with too little tumor tissue for whole panel sequencing.
In the current study tumor stage was distributed differently in some distinct mutations. EGFR mutations were found more often in patients with stage IV disease than in stage II, and stage I and II (18.0% vs. 10.3% vs. 12.0%, P=0.52). A similar observation was made in a recent study, showing that EGRF mutated patients significantly more often presented with distant metastases than wild type patients (14). This might be explained by the baseline characteristics of the EGFR cohorts. As these patients are younger and less likely to be smokers, decreased clinical suspicion of lung cancer may lead to delays in diagnosis. In addition to clinical factors, it is also possible that variations in tumor molecular biology influence invasive behavior and patterns of metastatic spread itself. This is supported by further differences in the distribution of driver mutations across stages. For instance, in contrast to EGFR mutated tumors.
PIK3CA and BRAF mutations were found to be more common in tumors diagnosed in earlier, more localized stages. Merged data from different cohorts and registers support our observation that EGFR mutations are more common in patients with metastatic disease and BRAF mutation are more frequent in early stages (15). A mutation in BRAF was also found using liquid biopsy in one patient with early-stage NSCLC in cohort of 20 patients, where a mutation would not have been expected due to the sample size (16). Targeted drugs for both BRAF and PIK3CA are available or in development (7,17,18). While only EGFR mutated NSCLC is eligible for adjuvant tyrosine kinase inhibitor (TKI) treatment at the moment, the encouraging data on osimertinib in the adjuvant setting (8) have paved the way for an array of adjuvant trials in other driver mutation indications. It is therefore reasonable to expect that panel testing, rather than sole EGFR testing, will become standard of care in resected NSCLC, and that targeted treatments will play an increasing role in the perioperative systemic treatment of lung cancer.
Comparing the mutational profile of non-squamous and squamous NSCLCs in our cohort it appeared, that non-squamous carcinomas are much more diverse with 23 different affected genes compared to 13 genes in the squamous group. Furthermore, we found that MET alterations occur more often in non-squamous histology (13.1% vs. 3.1%), whereas PIK3CA mutations have been found significantly more frequent in SCCs (2.5% vs. 21.2%). This is in line with previous reports, showing that SCCs harbor fewer oncogenic driver gene mutations compared to ACs (19). As in our analysis, this comprehensive study revealed also PIK3CA and TP53 to be significantly more frequent in SCCs (19). Interestingly, the proportion of EGFR mutations in the squamous cell group of our analysis is higher than reported in other cohorts. This might be due to a selection bias in this group with higher proportion of young and non-smoking patients.
In our cohort, we found no difference in OS between patients with or without mutations. This might be because some targetable mutations, like EGFR or ALK, improve the outcome as in these cases there is a possibility of targeted treatment. On the other hand, PIK3CA or KRAS mutations are associated with poorer prognosis. The number of patients in our cohort was too low to reasonably compare survival of single mutations.
Our study comprises some important limitations. First, the small number of patients and the retrospective character of the analysis does not allow extensive survival analyses. Additionally, a small sample size might not have enough power to detect all relevant differences, and a larger (public) dataset might be needed to analyze more precise gene alteration in lung cancer. However, we also believe that although using data from a single center might lead to a smaller dataset, a big advantage is that this data allows a comprehensive analysis of clinical features not available in public datasets. Second, especially in the squamous cell group, there might be a relevant selection bias, because panel testing in this cohort was not done in all patients but rather in selected squamous cell lung cancer patients only. Therefore, among the selected patients, females and never-smoker might be overrepresented and confound the results in this group. On the other hand, the unexpectedly high proportion of actionable mutations in the squamous cell group should serve to encourage molecular testing in non-AC NSCLC histologies, in particular in younger patients and never-smokers. Additionally, we are aware that not all results in our analysis are clinically relevant, and can help making treatment decisions. However, the aim in this study was to give a full overview of gene mutations in NSCLC patients with as many gene alterations as possible.
NGS panel testing is now routine in many countries for stage IV non-squamous NSCLC; however, registry data suggest that testing is not universally carried out and many patients are not tested for driver mutations, perhaps due to limited amount of material or lacking reimbursement for distinct histologies or localized stages (4). In the context of limited resources for testing outside stage IV, our results help prioritize patient for testing. In addition, our data clearly support the inclusion of squamous cell histologies in molecular testing, as oncogenic driver mutations can be detected regularly in this group, leading to additional treatment options. This is also supported by other recent publications showing that up to 85% of NSCLCs harbor a potentially actionable molecular alteration (16). It is important to avoid, that a patient is marked as “not mutated” only due to histology or due to negative results of sole EGFR and ALK testing.
Conclusions
Mutation profiles differed by histological type and stage and were significantly associated with PD-L1 expression. KRAS and EGFR mutations in SCC were more commonly found in our cohort than previously reported and PIK3CA revealed as oncogenic driver regularly found in SCC. In the context of limited resources, our results may help prioritize patient for testing when tissue material and funding is limited.
Acknowledgments
Parts of the research in this paper was presented at the 2022 ESMO conference in Prague as an electronic poster presentation. The poster’s abstract was published in the ESMO 2022 Abstract Book, a supplement to the official ESMO journal, Annals of Oncology: https://doi.org/10.1016/j.annonc.2022.02.218.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-751/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-751/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-751/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-751/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval for this study was obtained from the ethics board of the Ludwig-Maximilians University Munich (reference number 474-16 UE). Individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Kauffmann-Guerrero D, Kahnert K, Huber RM. Treatment Sequencing for Anaplastic Lymphoma Kinase-Rearranged Non-Small-Cell Lung Cancer. Drugs 2021;81:87-100. [Crossref] [PubMed]
- Griesinger F, Eberhardt W, Nusch A, et al. Biomarker testing in non-small cell lung cancer in routine care: Analysis of the first 3,717 patients in the German prospective, observational, nation-wide CRISP Registry (AIO-TRK-0315). Lung Cancer 2021;152:174-84. [Crossref] [PubMed]
- Chia PL, Mitchell P, Dobrovic A, et al. Prevalence and natural history of ALK positive non-small-cell lung cancer and the clinical impact of targeted therapy with ALK inhibitors. Clin Epidemiol 2014;6:423-32. [Crossref] [PubMed]
- Lohinai Z, Hoda MA, Fabian K, et al. Distinct Epidemiology and Clinical Consequence of Classic Versus Rare EGFR Mutations in Lung Adenocarcinoma. J Thorac Oncol 2015;10:738-46. [Crossref] [PubMed]
- Kauffmann-Guerrero D, Tufman A. Rare driver alterations in nonsmall cell lung cancer: novel targeted drugs. Curr Opin Oncol 2022;34:77-82. [Crossref] [PubMed]
- Wu YL, Tsuboi M, He J, et al. Osimertinib in Resected EGFR-Mutated Non-Small-Cell Lung Cancer. N Engl J Med 2020;383:1711-23. [Crossref] [PubMed]
- Sultova E, Westphalen CB, Jung A, et al. Implementation of Precision Oncology for Patients with Metastatic Breast Cancer in an Interdisciplinary MTB Setting. Diagnostics (Basel) 2021;11:733. [Crossref] [PubMed]
- Schoenfeld AJ, Rizvi H, Bandlamudi C, et al. Clinical and molecular correlates of PD-L1 expression in patients with lung adenocarcinomas. Ann Oncol 2020;31:599-608. [Crossref] [PubMed]
- Li C, Liu J, Xie Z, et al. PD-L1 expression with respect to driver mutations in non-small cell lung cancer in an Asian population: a large study of 1370 cases in China. Ther Adv Med Oncol 2020;12:1758835920965840. [Crossref] [PubMed]
- Zhang M, Li G, Wang Y, et al. PD-L1 expression in lung cancer and its correlation with driver mutations: a meta-analysis. Sci Rep 2017;7:10255. [Crossref] [PubMed]
- Liu J, Liu Y. Molecular diagnostic characteristics based on the next generation sequencing in lung cancer and its relationship with the expression of PD-L1. Pathol Res Pract 2020;216:152797. [Crossref] [PubMed]
- Grosse A, Grosse C, Rechsteiner M, et al. Analysis of the frequency of oncogenic driver mutations and correlation with clinicopathological characteristics in patients with lung adenocarcinoma from Northeastern Switzerland. Diagn Pathol 2019;14:18. [Crossref] [PubMed]
- Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer 2019;19:495-509. [Crossref] [PubMed]
- Mastromarino MG, Parini S, Azzolina D, et al. Liquid Biopsy Detecting Circulating Tumor Cells in Patients with Non-Small Cell Lung Cancer: Preliminary Results of a Pilot Study. Biomedicines 2023;11:153. [Crossref] [PubMed]
- Frank MS, Bodtger U, Gehl J, et al. Actionable Molecular Alterations Are Revealed in Majority of Advanced Non-Small Cell Lung Cancer Patients by Genomic Tumor Profiling at Progression after First Line Treatment. Cancers (Basel) 2021;14:132. [Crossref] [PubMed]
- Planchard D, Besse B, Groen HJM, et al. Phase 2 Study of Dabrafenib Plus Trametinib in Patients With BRAF V600E-Mutant Metastatic NSCLC: Updated 5-Year Survival Rates and Genomic Analysis. J Thorac Oncol 2022;17:103-15. [Crossref] [PubMed]
- Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. [Crossref] [PubMed]


